Gene transcriptions/RNA polymerase IIs
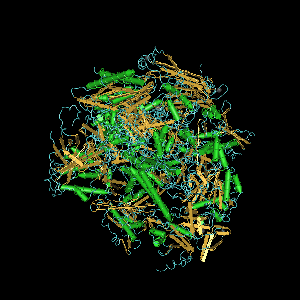
RNA polymerase II is a specific RNA polymerase that usually is the key part of the catalysis process that produces each RNA from the DNA gene or isoform to ultimately make a protein.
Subunits
[edit | edit source]The eukaryotic core RNA polymerase II was first purified using transcription assays.[2] The purified enzyme has typically 10-12 subunits (12 in humans and yeast) and is incapable of specific promoter recognition.[3] Many subunit-subunit interactions are known.[4]

Computer generated image of POLR2A gene colorized subunits: green - RPB1 domain 1, blue - RPB1 domain 2, sand - RPB1 domain 3, light blue - RPB1 domain 4, brown - RPB1 domain 6, and magenta - RPB1 CTD.
DNA-directed RNA polymerase II subunit RPB1 is an enzyme that in humans is encoded by the POLR2A gene. RPB1 is the largest subunit of RNA polymerase II. It contains a carboxy terminal domain (CTD) composed of heptapeptide repeats that are essential for polymerase activity. In combination with several other polymerase subunits, it forms the DNA binding domain of the polymerase, a groove in which the DNA template is transcribed into RNA.[5] It strongly interacts with RPB8.[4]
RPB2 (POLR2B) is the second largest subunit which in combination with at least two other polymerase subunits forms a structure within the polymerase that maintains contact in the active site of the enzyme between the DNA template and the newly synthesized RNA.[6]
The third largest subunit RPB3 (POLR2C) exists as a heterodimer with POLR2J forming a core subassembly. RPB3 strongly interacts with RPB1-5, 7, 10-12.[4]
RNA polymerase II subunit B4 (RPB4) encoded by the POLR2D gene[7] is the fourth largest subunit and may have a stress protective role.
In humans RPB5 is encoded by the POLR2E gene. Two molecules of this subunit are present in each RNA polymerase II.[8] RPB5 strongly interacts with RPB1, RPB3, and RPB6.[4]
RPB6 (POLR2F) forms a structure with at least two other subunits that stabilizes the transcribing polymerase on the DNA template.[9]
POLR2G encodes RPB7 that may play a role in regulating polymerase function.[10] RPB7 interacts strongly with RPB1 and RPB5.[4]
RPB8 (POLR2H) interacts with subunits RPB1-3, 5, and 7.[4]
The groove in which the DNA template is transcribed into RNA is composed of RPB9 (POLR2I) and RPB1.
RPB10 is the product of gene POLR2L. It interacts with RPB1-3 and 5, and strongly with RPB3.[4]
The RPB11 subunit is itself composed of three subunits in humans: POLR2J (RPB11-a), POLR2J2 (RPB11-b), and POLR2J3[11] (RPB11-c).
Also interacting with RPB3 is RPB12 (POLR2K).[4]
Assembly
[edit | edit source]RPB3 is involved in RNA polymerase II assembly.[12] A subcomplex of RPB2 and RPB3 appears soon after subunit synthesis.[12] This complex subsequently interacts with RPB1.[12]
RPB3, RPB5, and RPB7 interact with themselves to form homodimers, and RPB3 and RPB5 together are able to contact all of the other RPB subunits, except RPB9.[4] Only RPB1 strongly binds to RPB5.[4] The RPB1 subunit also contacts RPB7, RPB10, and more weakly but most efficiently with RPB8.[4] Once RPB1 enters the complex, other subunits such as RPB5 and RPB7 can enter, where RPB5 binds to RPB6 and RPB8 and RPB3 brings in RPB10, RPB 11, and RPB12.[4] RPB4 and RPB9 may enter once most of the complex is assembled. RPB4 forms a complex with RPB7.[4]
Kinetics
[edit | edit source]Enzymes can catalyze up to several million reactions per second. Enzyme rates depend on solution conditions and substrate concentration. Like other enzymes POLR2 has a saturation curve and a maximum velocity (Vmax). It has a Michaelis-Menten constant (Km) (substrate concentration required for one-half Vmax) and a kcat (the number of substrate molecules handled by one active site per second. The specificity constant is given by kcat/Km. The theoretical maximum for the specificity constant is the diffusion limit of about 108 to 109 (M-1 s-1), where every collision of the enzyme with its substrate results in catalysis.
The turnover number for RNA polymerase II is 0.16 s-1 subject to concentration.[13] RNA polymerase II switches between inactivated and activated states by translocating back and forth along the DNA.[14] Concentrations of [NTP]eq = 10 μM GTP, 10 μM UTP, 5 μM ATP and 2.5 μM CTP, produce a mean elongation rate, turnover number, of ~1 bp (NTP) s-1.[14]
Holoenzymes
[edit | edit source]RNA polymerase II holoenzyme is a form of eukaryotic RNA polymerase II that is recruited to the promoters of protein-coding genes in living cells.[3] It consists of RNA polymerase II, a subset of general transcription factors, and regulatory proteins known as SRB proteins.
Part of the assembly of the holoenzyme is referred to as the preinitiation complex, because its assembly takes place on the gene promoter before the initiation of transcription. The mediator complex acts as a bridge between RNA polymerase II and the transcription factors.
N-terminus
[edit | edit source]The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) refers to the start of a protein or polypeptide terminated by an amino acid with a free amine group (-NH2). The convention for writing peptide sequences is to put the N-terminus on the left and write the sequence from N- to C-terminus. When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus.
The N-terminus is the first part of the protein that exits the ribosome during protein biosynthesis. It often contains sequences that act as targeting signals, basically intracellular zip codes, that allow for the protein to be delivered to its designated location within the cell. The targeting signal is usually cleaved off after successful targeting by a processing peptidase. Some proteins are modified posttranslationally.
C-terminus
[edit | edit source]The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal end, or COOH-terminus) of a protein or polypeptide is the end of the amino acid chain terminated by a free carboxyl group (-COOH). The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus.
Each amino acid has a carboxyl group and an amine group, and amino acids link to one another to form a chain by a dehydration reaction by joining the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an unbound carboxyl group, the C-terminus, and an end with an amine group, the N-terminus. Proteins are naturally synthesized starting from the N-terminus and ending at the C-terminus.
The C-terminus can contain retention signals for protein sorting. The most common ER retention signal is the amino acid sequence -KDEL (or -HDEL) at the C-terminus, which keeps the protein in the endoplasmic reticulum and prevents it from entering the secretory pathway.
The C-terminus of proteins can be modified posttranslationally, for example, most commonly by the addition of a lipid anchor to the C-terminus that allows the protein to be inserted into a membrane without having a transmembrane domain. With Pol II, the C-terminus of RPB1 is appended to form the C-terminal domain (CTD).
Transcription initiation
[edit | edit source]The completed assembly of the holoenzyme with transcription factors and RNA polymerase II bound to the promoter forms the eukaryotic transcription initiation complex. Transcription in the archaea domain is similar to transcription in eukaryotes.[15]
Transcription begins with matching of NTPs to the first and second in the DNA sequence. This, like most of the remainder of transcription, is an energy-dependent process, consuming adenosine triphosphate (ATP) or other NTP.
Promoter clearance
[edit | edit source]After the first bond is synthesized, the RNA polymerase must clear the promoter. During this time there is a tendency to release the RNA transcript and produce truncated transcripts. This is called abortive initiation and is common for both eukaryotes and prokaroytes[16]. Abortive initiation continues to occur until the σ factor rearranges, resulting in the transcription elongation complex (which gives a 35 bp moving footprint). The σ factor is released before 80 nucleotides of mRNA are synthesized[17]. Once the transcript reaches approximately 23 nucleotides, it no longer slips and elongation can occur.
Human genes
[edit | edit source]Gene ID: 5430 is POLR2A RNA polymerase II subunit A. "This gene encodes the largest subunit of RNA polymerase II, the polymerase responsible for synthesizing messenger RNA in eukaryotes. The product of this gene contains a carboxy terminal domain composed of heptapeptide repeats that are essential for polymerase activity. These repeats contain serine and threonine residues that are phosphorylated in actively transcribing RNA polymerase. In addition, this subunit, in combination with several other polymerase subunits, forms the DNA binding domain of the polymerase, a groove in which the DNA template is transcribed into RNA."[18]
See also
[edit | edit source]References
[edit | edit source]- ↑ Meyer PA, Ye P, Zhang M, Suh MH, Fu J (Jun 2006). "Phasing RNA polymerase II using intrinsically bound Zn atoms: an updated structural model". Structure. 14 (6): 973-82. PMID 16765890. http://linkinghub.elsevier.com/retrieve/pii/S0969212606002152.
- ↑ Sawadogo M, Sentenac A (1990). Annu Rev Biochem. 59: 711-54.
- ↑ 3.0 3.1 Myer VE, Young RA (October 1998). "RNA polymerase II holoenzymes and subcomplexes". J. Biol. Chem. 273 (43): 27757–60. PMID 9774381. http://www.jbc.org/cgi/reprint/273/43/27757.pdf.
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 Acker J, de Graaff M, Cheynel I, Khazak V, Kedinger C, Vigneron M (Jul 1997). "Interactions between the human RNA polymerase II subunits". J Biol Chem. 272 (27): 16815-21. PMID 9201987.
- ↑ Entrez Gene: POLR2A polymerase (RNA) II (DNA directed) polypeptide A, 220kDa. http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5430.
- ↑ Entrez Gene: POLR2B polymerase (RNA) II (DNA directed) polypeptide B, 140kDa. http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5431.
- ↑ Khazak V, Estojak J, Cho H, Majors J, Sonoda G, Testa JR, Golemis EA (May 1998). "Analysis of the interaction of the novel RNA polymerase II (pol II) subunit hsRPB4 with its partner hsRPB7 and with pol II". Mol Cell Biol. 18 (4): 1935-45. PMID 9528765. PMC 121423. //www.ncbi.nlm.nih.gov/pmc/articles/PMC121423/.
- ↑ Entrez Gene: POLR2E polymerase (RNA) II (DNA directed) polypeptide E, 25kDa. http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5434.
- ↑ Entrez Gene: POLR2F polymerase (RNA) II (DNA directed) polypeptide F. http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5435.
- ↑ Entrez Gene: POLR2G polymerase (RNA) II (DNA directed) polypeptide G. http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5436.
- ↑ POLR2J3 polymerase (RNA) II (DNA directed) polypeptide J3. http://www.ncbi.nlm.nih.gov/gene/548644?ordinalpos=1&itool=EntrezSystem2.PEntrez.Gene.Gene_ResultsPanel.Gene_RVDocSum.
- ↑ 12.0 12.1 12.2 Kolodziej PA, Young RA (Sep 1991). "Mutations in the three largest subunits of yeast RNA polymerase II that affect enzyme assembly". Mol Cell Biol. 11 (9): 4669-78. http://mcb.asm.org/cgi/reprint/11/9/4669?ijkey=9d60d05ed32981de57ecc990796689311e8f86a0.
- ↑ Jin J, Dong W, Guarino LA (Dec 1998). "The LEF-4 subunit of Baculovirus RNA polymerase has RNA 5'-triphosphatase and ATPase activities". J Virol. 72 (12): 10011-9. http://jvi.highwire.org/cgi/reprint/72/12/10011.
- ↑ 14.0 14.1 Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM (Nov 2005). "Direct observation of base-pair stepping by RNA polymerase". Nature. 438 (7067): 460-5. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1356566/.
- ↑ Ouhammouch M, Dewhurst RE, Hausner W, Thomm M, Geiduschek EP (2003). "Activation of archaeal transcription by recruitment of the TATA-binding protein". Proc Nat Acad Sci (USA). 100 (9): 5097–102. doi:10.1073/pnas.0837150100. PMID 12692306. PMC 154304. //www.ncbi.nlm.nih.gov/pmc/articles/PMC154304/.
- ↑ . doi:10.1126/science.1169237.
- ↑ . PMID 12213653.
- ↑ RefSeq (July 2008). "POLR2A RNA polymerase II subunit A [ Homo sapiens (human) ]". Bethesda, MD, USA: National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved 20 January 2020.
