Gene transcriptions/Boxes/Enhancers

"An E-box (Enhancer Box) is a DNA sequence which usually lies upstream of a gene in a promoter region."[1]
Enhancers
[edit | edit source]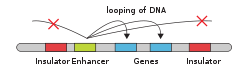
"An enhancer is a short region of DNA that can be bound with proteins (namely, the trans-acting factors, much like a set of transcription factors) to enhance transcription levels of genes (hence the name) in a gene cluster. While enhancers are usually cis-acting, an enhancer does not need to be particularly close to the genes it acts on, and sometimes need not be located on the same chromosome.[2]
In eukaryotic cells the structure of the chromatin complex of DNA is folded in a way that although the enhancer DNA is far from the gene in regard to the number of nucleotides, it is geometrically close to the promoter and gene.
An enhancer may be located upstream or downstream of the gene it regulates.
Enhancers do not act on the promoter region itself, but are bound by activator proteins. These activator proteins interact with the mediator complex, which recruits polymerase II and the general transcription factors which then begin transcribing the genes. Enhancers can also be found within introns. An enhancer's orientation may even be reversed without affecting its function. Additionally, an enhancer may be excised and inserted elsewhere in the chromosome, and still affect gene transcription.
Def. a "short region of DNA that can increase transcription of genes"[3] is called an enhancer.
Boxes
[edit | edit source]A "repeating sequence of nucleotides that forms a transcription or a regulatory signal"[4] is a box.
Immunoglobulin domains
[edit | edit source]The immunoglobulin domain is a type of protein domain that consists of a 2-layer sandwich of between 7 and 9 antiparallel β-strands arranged in two β-sheets with a Greek key topology.[5][6]
The E-box is a control element in immunoglobulin heavy-chain promoters.[7]
Consensus sequences
[edit | edit source]The consensus sequence for the E-box element is CANNTG, with a palindromic canonical sequence of CACGTG.[8]
Proximal promoters
[edit | edit source]"[T]he proximal sequence upstream of the gene that tends to contain primary regulatory elements" is a proximal promoter.[9]
It is "[a]pproximately 250 base pairs [or nucleotides, nts] upstream of the [transcription] start site".[9]
There may be an E box in the proximal promoter of some genes.[8]
Distal promoters
[edit | edit source]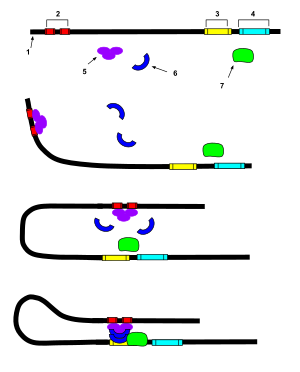
An E-box usually lies within the distal promoter starting at or near -300 nts, the proximal promoter, or both.[8]
Hypotheses
[edit | edit source]- A1BG is not transcribed by an enhancer box.
- Existence of an enhancer box on either side of A1BG does not prove that it is actively used to transcribe A1BG.
- A1BG is not transcribed by a downstream enhancer box.
Samplings
[edit | edit source]Regarding hypotheses 1:
A1BG has four possible transcription directions:
- on the negative strand from ZSCAN22 to A1BG,
- on the positive strand from ZSCAN22 to A1BG,
- on the negative strand from ZNF497 to A1BG, and
- on the positive strand from ZNF497 to A1BG.
For each transcription promoter that interacts directly with RNA polymerase II holoenzyme, the four possible consensus sequences need to be tested on the four possible transcription directions, even though some genes may only be transcribed from the negative strand in the 3'-direction on the transcribed strand.
For the Basic programs (starting with SuccessablesE.bas) written to compare nucleotide sequences with the sequences on either the template strand (-), or coding strand (+), of the DNA, in the negative direction (-), or the positive direction (+), the programs are looking for, and found:
- negative strand in the negative direction is SuccessablesE--.bas, looking for CA(A/C/G/T)(A/C/G/T)TG, 10, CAGATG at 4212, CATTTG at 3482, CAGATG at 2988, CACCTG at 2116, CAGTTG at 1513, CAGATG at 1224, CAAGTG at 1179, CACATG at 797, CAGATG at 481, CACATG at 324.
- negative strand in the positive direction is SuccessablesE-+.bas, looking for 3'-C-A-(A/C/G/T)-(A/C/G/T)-T-G-5', 26, 3'-CAGGTG-5', 196, 3'-CACGTG-5', 570, 3'-CACCTG-5', 858, 3'-CACCTG-5', 958, 3'-CAGGTG-5', 1968, 3'-CACATG-5', 2031, 3'-CAGCTG-5', 2054, 3'-CAGGTG-5', 2127, 3'-CACCTG-5', 2249, 3'-CAGGTG-5', 2374, 3'-CAGCTG-5', 2404, 3'-CACCTG-5', 2432, 3'-CAAGTG-5', 2510, 3'-CACCTG-5', 2568, 3'-CACCTG-5', 3046, 3'-CAGGTG-5', 3149, 3'-CAGCTG-5', 3241, 3'-CATCTG-5', 3404, 3'-CAGATG-5', 3475, 3'-CACATG-5', 3707, 3'-CACATG-5', 3742, 3'-CAGCTG-5', 3777, 3'-CATGTG-5', 3902, 3'-CACATG-5', 3956, 3'-CATGTG-5', 3958, 3'-CACTTG-5', 4015,
- positive strand in the negative direction is SuccessablesE+-.bas, looking for 3'-C-A-(A/C/G/T)-(A/C/G/T)-T-G-5', 21, 3'-CATATG-5', 41, 3'-CATTTG-5', 364, 3'-CACCTG-5', 393, 3'-CACCTG-5', 1130, 3'-CACCTG-5', 1172, 3'-CAAATG-5', 1579, 3'-CAGGTG-5', 2079, 3'-CACTTG-5', 2126, 3'-CAGGTG-5', 2570, 3'-CACTTG-5', 2579, 3'-CACATG-5', 2667, 3'-CACTTG-5', 2920, 3'-CACTTG-5', 3102, 3'-CACTTG-5', 3241, 3'-CAGATG-5', 3620, 3'-CAGATG-5', 3627, 3'-CAACTG-5', 3850, 3'-CAGATG-5', 3919, 3'-CAGGTG-5', 3953, 3'-CACCTG-5', 3969, 3'-CACTTG-5', 4011,
- positive strand in the positive direction is SuccessablesE++.bas, looking for 3'-C-A-(A/C/G/T)-(A/C/G/T)-T-G-5', 11, 3'-CACCTG-5', 186, 3'-CACGTG-5', 547, 3'-CATGTG-5', 567, 3'-CACGTG-5', 1219, 3'-CAGGTG-5', 1843, 3'-CAGGTG-5', 2028, 3'-CACGTG-5', 2961, 3'-CAGGTG-5', 3086, 3'-CACGTG-5', 3884, 3'-CACTTG-5', 3936, 3'-CAAGTG-5', 4202,
- complement, negative strand, negative direction is SuccessablesEc--.bas, looking for 3'-G-T-(A/C/G/T)-(A/C/G/T)-A-C-5', 21, 3'-GTATAC-5', 41, 3'-GTAAAC-5', 364, 3'-GTGGAC-5', 393, 3'-GTGGAC-5', 1130, 3'-GTGGAC-5', 1172, 3'-GTTTAC-5', 1579, 3'-GTCCAC-5', 2079, 3'-GTGAAC-5', 2126, 3'-GTCCAC-5', 2570, 3'-GTGAAC-5', 2579, 3'-GTGTAC-5', 2667, 3'-GTGAAC-5', 2920, 3'-GTGAAC-5', 3102, 3'-GTGAAC-5', 3241, 3'-GTCTAC-5', 3620, 3'-GTCTAC-5', 3627, 3'-GTTGAC-5', 3850, 3'-GTCTAC-5', 3919, 3'-GTCCAC-5', 3953, 3'-GTGGAC-5', 3969, 3'-GTGAAC-5', 4011,
- complement, negative strand, positive direction is SuccessablesEc-+.bas, looking for 3'-G-T-(A/C/G/T)-(A/C/G/T)-A-C-5', 11, 3'-GTGGAC-5', 186, 3'-GTGCAC-5', 547, 3'-GTACAC-5', 567, 3'-GTGCAC-5', 1219, 3'-GTCCAC-5', 1843, 3'-GTCCAC-5', 2028, 3'-GTGCAC-5', 2961, 3'-GTCCAC-5', 3086, 3'-GTGCAC-5', 3884, 3'-GTGAAC-5', 3936, 3'-GTTCAC-5', 4202,
- complement, positive strand, negative direction is SuccessablesEc+-.bas, looking for 3'-G-T-(A/C/G/T)-(A/C/G/T)-A-C-5', 10 , 3'-GTGTAC-5', 324 , 3'-GTCTAC-5', 481 , 3'-GTGTAC-5', 797 , 3'-GTTCAC-5', 1179 , 3'-GTCTAC-5', 1224 , 3'-GTCAAC-5', 1513 , 3'-GTGGAC-5', 2116 , 3'-GTCTAC-5', 2988 , 3'-GTAAAC-5', 3482 , 3'-GTCTAC-5', 4212,
- complement, positive strand, positive direction is SuccessablesEc++.bas, looking for 3'-G-T-(A/C/G/T)-(A/C/G/T)-A-C-5', 26, 3'-GTCCAC-5', 196, 3'-GTGCAC-5', 570, 3'-GTGGAC-5', 858, 3'-GTGGAC-5', 958, 3'-GTCCAC-5', 1968, 3'-GTGTAC-5', 2031, 3'-GTCGAC-5', 2054, 3'-GTCCAC-5', 2127, 3'-GTGGAC-5', 2249, 3'-GTCCAC-5', 2374, 3'-GTCGAC-5', 2404, 3'-GTGGAC-5', 2432, 3'-GTTCAC-5', 2510, 3'-GTGGAC-5', 2568, 3'-GTGGAC-5', 3046, 3'-GTCCAC-5', 3149, 3'-GTCGAC-5', 3241, 3'-GTAGAC-5', 3404, 3'-GTCTAC-5', 3475, 3'-GTGTAC-5', 3707, 3'-GTGTAC-5', 3742, 3'-GTCGAC-5', 3777, 3'-GTACAC-5', 3902, 3'-GTGTAC-5', 3956, 3'-GTACAC-5', 3958, 3'-GTGAAC-5', 4015.
The complement inverse is the same as the direct consensus sequence.
Enhancer box UTRs
[edit | edit source]Negative strand, negative direction: CAGATG at 4212, CATTTG at 3482, CAGATG at 2988.
Positive strand, negative direction: CACTTG at 4011, CACCTG at 3969, CAGGTG at 3953, CAGATG at 3919, CAACTG at 3850, CAGATG at 3627, CAGATG at 3620, CACTTG at 3241, CACTTG at 3102, CACTTG at 2920.
Enhancer box proximal promoters
[edit | edit source]Positive strand, negative direction: CACATG at 2667.
Positive strand, positive direction: CAAGTG at 4202.
Enhancer box distal promoters
[edit | edit source]Negative strand, negative direction: CACCTG at 2116, CAGTTG at 1513, CAGATG at 1224, CAAGTG at 1179, CACATG at 797, CAGATG at 481, CACATG at 324.
Positive strand, negative direction: CACTTG at 2579, CAGGTG at 2570, CACTTG at 2126, CAGGTG at 2079, CAAATG at 1579, CACCTG at 1172, CACCTG at 1130, CACCTG at 393, CATTTG at 364, CATATG at 41.
Negative strand, positive direction: CACTTG at 4015, CATGTG at 3958, CACATG at 3956, CATGTG at 3902, CAGCTG at 3777, CACATG at 3742, CACATG at 3707, CAGATG at 3475, CATCTG at 3404, CAGCTG at 3241, CAGGTG at 3149, CACCTG at 3046, CACCTG at 2568, CAAGTG at 2510, CACCTG at 2432, CAGCTG at 2404, CAGGTG at 2374, CACCTG at 2249, CAGGTG at 2127, CAGCTG at 2054, CACATG at 2031, CAGGTG at 1968, CACCTG at 958, CACCTG at 858, CACGTG at 570, CAGGTG at 196.
Positive strand, positive direction: CACTTG at 3936, CACGTG at 3884, CAGGTG at 3086, CACGTG at 2961, CAGGTG at 2028, CAGGTG at 1843, CACGTG at 1219, CATGTG at 567, CACGTG at 547, CACCTG at 186.
Enhancer box random dataset samplings
[edit | edit source]- Er0: 12, CACGTG at 4343, CATGTG at 3956, CAATTG at 3880, CAACTG at 3533, CAACTG at 3467, CAGTTG at 3440, CAGGTG at 3398, CAATTG at 3202, CAATTG at 2233, CATATG at 2151, CATGTG at 1059, CACCTG at 999.
- Er1: 10, CAATTG at 4110, CATTTG at 4051, CATGTG at 3891, CAGTTG at 3388, CAAATG at 3372, CAACTG at 2752, CATATG at 2101, CATATG at 1605, CATCTG at 1131, CAAATG at 263.
- Er2: 15, CAGTTG at 4536, CAGCTG at 4212, CAAATG at 3829, CACATG at 3734, CAGTTG at 3245, CATTTG at 2627, CAAGTG at 2604, CAGGTG at 2198, CACCTG at 2124, CAGTTG at 1987, CAGATG at 1826, CATATG at 1757, CAAATG at 427, CATCTG at 203, CATGTG at 166.
- Er3: 13, CACGTG at 3769, CAGGTG at 3527, CATCTG at 3286, CAAGTG at 3239, CAGGTG at 2880, CACTTG at 2805, CATCTG at 1770, CATGTG at 1134, CAAATG at 1055, CAGATG at 298, CAAGTG at 266, CAAATG at 158, CATGTG at 93.
- Er4: 18, CAGTTG at 4419, CAGATG at 4202, CATTTG at 2905, CATCTG at 2584, CACGTG at 2287, CATGTG at 2243, CATGTG at 2224, CACCTG at 1958, CACCTG at 1913, CAAATG at 1809, CATATG at 1685, CAGTTG at 1578, CAAATG at 1363, CACTTG at 1162, CACCTG at 1123, CACATG at 909, CATTTG at 836, CACTTG at 646.
- Er5: 12, CACTTG at 3937, CACCTG at 3116, CATTTG at 2790, CAATTG at 2227, CAGGTG at 2213, CAGCTG at 2162, CATATG at 1779, CAATTG at 1579, CATTTG at 1204, CAACTG at 1180, CAGGTG at 697, CACGTG at 59.
- Er6: 16, CACCTG at 4358, CAACTG at 3821, CAGGTG at 3800, CAGTTG at 3329, CAAATG at 3078, CACGTG at 2905, CAGCTG at 2881, CAGTTG at 2638, CACATG at 2601, CAATTG at 2488, CATGTG at 2330, CACTTG at 2062, CAACTG at 1809, CATGTG at 1342, CACGTG at 654, CATATG at 245.
- Er7: 11, CATTTG at 4381, CACTTG at 3970, CACTTG at 3118, CAAATG at 3034, CATTTG at 2446, CAATTG at 2357, CACGTG at 1856, CACCTG at 1452, CATTTG at 1177, CATCTG at 1159, CAACTG at 20.
- Er8: 12, CACTTG at 4159, CAGATG at 3821, CAACTG at 3658, CAGATG at 2726, CATTTG at 2428, CAGTTG at 2300, CATCTG at 1711, CAAATG at 1376, CACTTG at 1254, CAAATG at 963, CAGGTG at 292, CAAATG at 213.
- Er9: 14, CAGATG at 4485, CACATG at 4071, CAGATG at 4027, CACTTG at 3646, CATTTG at 3619, CATTTG at 3477, CAAATG at 2077, CATGTG at 1975, CACCTG at 1898, CAACTG at 1697, CATTTG at 1448, CACGTG at 1187, CACCTG at 922, CAACTG at 121.
Er UTRs
[edit | edit source]- Er0: CACGTG at 4343, CATGTG at 3956, CAATTG at 3880, CAACTG at 3533, CAACTG at 3467, CAGTTG at 3440, CAGGTG at 3398, CAATTG at 3202.
- Er2: CAGTTG at 4536, CAGCTG at 4212, CAAATG at 3829, CACATG at 3734, CAGTTG at 3245.
- Er4: CAGTTG at 4419, CAGATG at 4202, CATTTG at 2905.
- Er6: CACCTG at 4358, CAACTG at 3821, CAGGTG at 3800, CAGTTG at 3329, CAAATG at 3078, CACGTG at 2905, CAGCTG at 2881.
- Er8: CACTTG at 4159, CAGATG at 3821, CAACTG at 3658.
Er core promoters
[edit | edit source]- Er7: CATTTG at 4381.
- Er9: CAGATG at 4485.
Er proximal promoters
[edit | edit source]- Er2: CATTTG at 2627, CAAGTG at 2604.
- Er6: CAGTTG at 2638, CACATG at 2601.
- Er8: CAGATG at 2726.
- Er1: CAATTG at 4110, CATTTG at 4051.
- Er9: CACATG at 4071.
Er distal promoters
[edit | edit source]- Er0: CAATTG at 2233, CATATG at 2151, CATGTG at 1059, CACCTG at 999.
- Er2: CAGGTG at 2198, CACCTG at 2124, CAGTTG at 1987, CAGATG at 1826, CATATG at 1757, CAAATG at 427, CATCTG at 203, CATGTG at 166.
- Er4: CATCTG at 2584, CACGTG at 2287, CATGTG at 2243, CATGTG at 2224, CACCTG at 1958, CACCTG at 1913, CAAATG at 1809, CATATG at 1685, CAGTTG at 1578, CAAATG at 1363, CACTTG at 1162, CACCTG at 1123, CACATG at 909, CATTTG at 836, CACTTG at 646.
- Er6: CAATTG at 2488, CATGTG at 2330, CACTTG at 2062, CAACTG at 1809, CATGTG at 1342, CACGTG at 654, CATATG at 245.
- Er8: CATTTG at 2428, CAGTTG at 2300, CATCTG at 1711, CAAATG at 1376, CACTTG at 1254, CAAATG at 963, CAGGTG at 292, CAAATG at 213.
- Er1: CATGTG at 3891, CAGTTG at 3388, CAAATG at 3372, CAACTG at 2752, CATATG at 2101, CATATG at 1605, CATCTG at 1131, CAAATG at 263.
- Er3: CACGTG at 3769, CAGGTG at 3527, CATCTG at 3286, CAAGTG at 3239, CAGGTG at 2880, CACTTG at 2805, CATCTG at 1770, CATGTG at 1134, CAAATG at 1055, CAGATG at 298, CAAGTG at 266, CAAATG at 158, CATGTG at 93.
- Er5: CACTTG at 3937, CACCTG at 3116, CATTTG at 2790, CAATTG at 2227, CAGGTG at 2213, CAGCTG at 2162, CATATG at 1779, CAATTG at 1579, CATTTG at 1204, CAACTG at 1180, CAGGTG at 697, CACGTG at 59.
- Er7: CACTTG at 3970, CACTTG at 3118, CAAATG at 3034, CATTTG at 2446, CAATTG at 2357, CACGTG at 1856, CACCTG at 1452, CATTTG at 1177, CATCTG at 1159, CAACTG at 20.
- Er9: CAGATG at 4027, CACTTG at 3646, CATTTG at 3619, CATTTG at 3477, CAAATG at 2077, CATGTG at 1975, CACCTG at 1898, CAACTG at 1697, CATTTG at 1448, CACGTG at 1187, CACCTG at 922, CAACTG at 121.
Response element analysis and results
[edit | edit source]Transcribed enhancer boxes
[edit | edit source]"MYC is a basic helix-loop-helix transcription factor, evolutionarily conserved in all vertebrates with a considerable amount of sequence similarity (Atchley & Fitch, 1995). It binds to thousands of promoters in mammalian cells as MYC-MAX heterodimer (Blackwood & Eisenman, 1991; C. Y. Lin et al., 2012). In particular it binds the motif CACGTG of the enhancer box (E-box) in the core promoter of active genes. Depending on the target gene, MYC can act as transcriptional activator or repressor, and, can affect transcription at both initiation and elongation steps (Rahl et al., 2010)."[10]
"MYC mediates the transcriptional response of growth-factors stimulation. Importantly, MYC does not only regulate the expression of mRNA(s), it also regulates ribosomal and tRNA genes, transcribed by the RNA Pol I and RNA Pol III respectively (Campbell & White, 2014; Dai, Sun, & Lu, 2010; Mitchell et al., 2015). Amongst the major gene ontology categories of protein-coding genes under the control of MYC there are: ribosome biogenesis, apoptosis, cell adhesion, cell size, angiogenesis and metabolic pathways (Nieminen, Partanen, & Klefstrom, 2007; Peterson & Ayer, 2011; A. M. Singh & Dalton, 2009; Uslu et al., 2014; van Riggelen, Yetil, & Felsher, 2010)."[10]
"The ATA box [AAATAT], GC box [GGCGGG], CArG box [CCTATTATGCG], [two E boxes CAGTTG] and M-CAT [CATTCCT] consensus sequences are [described from the mouse dystrophin promoter]."[11]
"The E box [ enhancer box ] sites that are most important are those of the E2 box class (GCAGXTGG/T). Two E2 box sites are present in the immunoglobulin heavy chain gene enhancer [...] and one is present in the kappa enhancer, designated KE2 [29-31]."[12]
"The developmental regulation of Ig gene expression is dependent on various sequences in the Ig enhancer. One class of such sequence elements is the E boxes. They share as a consensus sequence NNCANNTGNN. The E-box sites were first identified by dimethylsulfate protection experiments (6, 12). Factors were found to protect certain sequences from methylation in the Ig heavy- and light-chain enhancer in B cells but not in non-B cells (6,12). That the E-box elements are critical for B-cell-specific gene expression became evident from mutational analysis. Mutation of E-box sites caused a significant decrease in Ig transcription (18, 21). The most dramatic impact on Ig expression was found in mutations of elements that contain an E2 box (G/ACAGNTGT/G) (21). The E2 boxes are particularly interesting because they are also present in muscle-and pancreas-specific enhancers (3,4,32). Mutation of the E2-box elements present in these enhancers revealed the crucial role of these elements in regulating muscle- and pancreas-specific genes (16, 22, 26, 27, 32)."[13]
"The two E2 boxes in the mouse and human E-cadherin promoter sequences were demonstrated to play a crucial role in the epithelial-specific expression of E-cadherin Behrens et al. 1991, Giroldi et al. 1997. Mutation of these sequence elements results in upregulation of the E-cadherin promoter in dedifferentiated cancer cells, whereas the wild-type promoter shows low activity in such cells. Recently, it was shown that the zinc finger transcriptional repressor Snail can downregulate E-cadherin by binding to the E boxes in the E-cadherin promoter Batlle et al. 2000, Cano et al. 2000. Human Snail belongs to a family of zinc finger proteins, which contain four or five zinc finger domains of the C2H2 type at their C-terminal end. These zinc fingers bind to the CANNTG sequence in E box motifs."[14]
The CArG boxes occur between -400 and -200 nts, between the E boxes and the TCE element.[15]
The "isolated mouse chromogranin B promoter [specifically] the proximal chromogranin B promoter (from −216 to −91 bp); [...] contains an E box (at [−206 bp]CACCTG[−201 bp]), four G/C-rich regions (at[− 196 bp]CCCCGC[−191 bp], [−134 bp]CCGCCCGC[−127 bp],[− 125 bp]GGCGCCGCC[−117 bp], and [−115 bp]CGGGGC[−110 bp]), and a cAMP response element (CRE; at [−102 bp]TGACGTCA[−95 bp]). A 60-bp core promoter region, defined by an internal deletion from −134 to −74 bp upstream of the cap site and spanning the CRE and three G/C-rich regions, directed tissue-specific expression of the gene. The CRE motif directed cell type-specific expression of the chromogranin B gene in neurons, whereas three of the G/C-rich regions played a crucial role in neuroendocrine cells. Both the endogenous chromogranin B gene and the transfected chromogranin B promoter were induced by preganglionic secretory stimuli (pituitary adenylyl cyclase-activating polypeptide, vasoactive intestinal peptide, or a nicotinic cholinergic agonist), establishing stimulus-transcription coupling for this promoter. The adenylyl cyclase activator forskolin, nerve growth factor, and retinoic acid also activated the chromogranin B gene. Secretagogue-inducible expression of chromogranin B also mapped onto the proximal promoter; inducible expression was entirely lost upon internal deletion of the 60-bp core (from −134 to −74 bp). [...] CRE and G/C-rich domains are crucial determinants of both cell type-specific and secretagogue-inducible expression of the chromogranin B gene."[16]
See also
[edit | edit source]References
[edit | edit source]- ↑ "E-box". San Francisco, California: Wikimedia Foundation, Inc. April 13, 2013. Retrieved 2013-04-17.
- ↑ Charalampos G. Spilianakis, Maria D. Lalioti, Terrence Town, Gap Ryol Lee, Richard A. Flavell (2005). "Interchromosomal associations between alternatively expressed loci". Nature 435 (7042): 637–45. doi:10.1038/nature03574. PMID 15880101.
- ↑ SemperBlotto (16 January 2011). "enhancer". San Francisco, California: Wikimedia Foundation, Inc. Retrieved 2017-08-25.
{{cite web}}:|author=has generic name (help) - ↑ "Box (disambiguation)". San Francisco, California: Wikimedia Foundation, Inc. May 23, 2013. Retrieved 2013-06-15.
- ↑ Bork P, Holm L, Sander C (September 1994). "The immunoglobulin fold. Structural classification, sequence patterns and common core". Journal of Molecular Biology 242 (4): 309–20. doi:10.1006/jmbi.1994.1582. PMID 7932691.
- ↑ Brümmendorf T, Rathjen FG (1995). "Cell adhesion molecules 1: immunoglobulin superfamily". Protein Profile 2 (9): 963–1108. PMID 8574878.
- ↑ George M. Church, Anne Ephrussi, Walter Gilbert, Susumu Tonegawa (1985). Nature 313 (6005): 798-801.
- ↑ 8.0 8.1 8.2 Jaideep Chaudhary and Michael K. Skinner (May 1999). "Basic Helix-Loop-Helix Proteins Can Act at the E-Box within the Serum Response Element of the c-fos Promoter to Influence Hormone-Induced Promoter Activation in Sertoli Cells". Molecular Endocrinology 13 (5): 774-86. doi:10.1210/me.13.5.774. PMID 10319327. http://mend.endojournals.org/content/13/5/774.short. Retrieved 2013-06-14.
- ↑ 9.0 9.1 "Promoter (genetics)". San Francisco, California: Wikimedia Foundation, Inc. June 14, 2013. Retrieved 2013-06-15.
- ↑ 10.0 10.1 Massimo Petretich (20 September 2016). Importance of Chromosomal Architecture to Organize Promoter-Enhancer Long-Range Interactions in c-Myc locus. Heidelberg, Germany: Ruperto-Carola University of Heidelberg. pp. 195. http://archiv.ub.uni-heidelberg.de/volltextserver/21976/1/Thesis_Massimo_Petretich.pdf. Retrieved 2017-09-05.
- ↑ Shigemi Kimura, Kuniya Abe, Misao Suzuki, Masakatsu Ogawa, Kowashi Yoshioka, Tadasi Kaname, Teruhisa Miike, Ken‐ichi Yamamura (June 1997). "A 900 bp genomic region from the mouse dystrophin promoter directs lacZ reporter expression only to the right heart of transgenic mice". Development, Growth & Differentiation 39 (1): 257-265. doi:10.1046/j.1440-169X.1997.t01-2-00001.x. https://onlinelibrary.wiley.com/doi/pdf/10.1046/j.1440-169X.1997.t01-2-00001.x. Retrieved 25 March 2019.
- ↑ Cornelis Murre, Gretchen Bain, Marc A. van Dijk, Isaac Engel, Beth A. Furnari, Mark E. Massari, James R. Matthews, Melanie W. Quong, Richard R. Rivera, Maarten H. Stuiver (June 1994). "Structure and function of helix-loop-helix proteins". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression 1218 (2): 129-35. http://www.sciencedirect.com/science/article/pii/0167478194900019. Retrieved 2017-02-08.
- ↑ Gretchen Bain, Stefan Gruenwald, and Cornelis Murre (June 1993). "E2A and E2-2 are subunits of B-cell-specific E2-box DNA-binding proteins". Molecular and Cellular Biology 13 (6): 3522-3529. doi:10.1128/MCB.13.6.3522. http://mcb.asm.org/content/13/6/3522.full.pdf. Retrieved 2 February 2019.
- ↑ Joke Comijn, Geert Berx, Petra Vermassen, Kristin Verschueren, Leo van Grunsven, Erik Bruyneel, Marc Mareel, Danny Huylebroeck, Frans van Roy (June 2001). "The Two-Handed E Box Binding Zinc Finger Protein SIP1 Downregulates E-Cadherin and Induces Invasion". Molecular Cell 7 (6): 1267-78. doi:10.1016/S1097-2765(01)00260-X. https://www.sciencedirect.com/science/article/pii/S109727650100260X. Retrieved 11 January 2019.
- ↑ Oliver G. McDonald, Brian R. Wamhoff, Mark H. Hoofnagle, and Gary K. Owens (January 4, 2006). "Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo". The Journal of Clinical Investigation 116 (1): 36-48. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1323266/. Retrieved 2014-06-05.
- ↑ Nitish R. Mahapatra, Manjula Mahata, Arun K. Datta, Hans-Hermann Gerdes, Wieland B. Huttner, Daniel T. O’Connor, Sushil K. Mahata (1 October 2000). "Neuroendocrine Cell Type-Specific and Inducible Expression of the Chromogranin B Gene: Crucial Role of the Proximal Promoter". Endocrinology 141 (10): 3668–3678. doi:10.1210/endo.141.10.7725. https://academic.oup.com/endo/article/141/10/3668/2987577. Retrieved 15 September 2018.
Further reading
[edit | edit source]- Jaideep Chaudhary and Michael K. Skinner (May 1999). "Basic Helix-Loop-Helix Proteins Can Act at the E-Box within the Serum Response Element of the c-fos Promoter to Influence Hormone-Induced Promoter Activation in Sertoli Cells". Molecular Endocrinology 13 (5): 774-86. doi:10.1210/me.13.5.774. PMID 10319327. http://mend.endojournals.org/content/13/5/774.short. Retrieved 2013-06-14.
External links
[edit | edit source]- GenomeNet KEGG database
- Google scholar Advanced Scholar Search
- Home - Gene - NCBI
- NCBI All Databases Search
- NCBI Site Search
- Office of Scientific & Technical Information
- PubChem Public Chemical Database
- Scirus for scientific information only advanced search
- Enhancer box gene transcriptions
