WikiJournal of Medicine/Eukaryotic and prokaryotic gene structure
WikiJournal of Medicine
Open access • Publication charge free • Public peer review • Wikipedia-integrated
This article has been through public peer review.
Post-publication review comments or direct edits can be left at the version as it appears on Wikipedia.First submitted:
Accepted:
Reviewer comments
PDF: Download
DOI: 10.15347/wjm/2017.002
QID: Q28867140
XML: Download
Share article
![]() Email
|
Email
| ![]() Facebook
|
Facebook
| ![]() Twitter
|
Twitter
| ![]() LinkedIn
|
LinkedIn
| ![]() Mendeley
|
Mendeley
| ![]() ResearchGate
ResearchGate
Suggested citation format:
Thomas Shafee; Rohan Lowe (17 January 2017). "Eukaryotic and prokaryotic gene structure". WikiJournal of Medicine 4 (1). doi:10.15347/WJM/2017.002. Wikidata Q28867140. ISSN 2002-4436. https://upload.wikimedia.org/wikiversity/en/0/0c/Eukaryotic_and_prokaryotic_gene_structure.pdf.
Citation metrics
AltMetrics
Page views on Wikipedia
Wikipedia: Content from this work is used in the following Wikipedia article: Gene structure, Gene, Post-transcriptional modification, Regulatory sequence, Operon.
License: ![]()
![]() This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction, provided the original author and source are credited.
This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction, provided the original author and source are credited.
Michaël R. Laurent ![]() (handling editor) contact
(handling editor) contact
Robert Root-Bernstein ![]()
Alexandre P Y Lopes ![]()
Article information
Abstract
Introduction
Gene structure
Genes contain the information necessary for living cells to survive and reproduce.[1][2] In most organisms, genes are made of DNA, where the particular DNA sequence determines the function of the gene. A gene is transcribed (copied) from DNA into RNA, which can either be non-coding (ncRNA) with a direct function, or an intermediate messenger (mRNA) that is then translated into protein. Each of these steps is controlled by specific sequence elements, or regions, within the gene. Every gene, therefore, requires multiple sequence elements to be functional.[2] This includes the sequence that actually encodes the functional protein or ncRNA, as well as multiple regulatory sequence regions. These regions may be as short as a few base pairs, up to many thousands of base pairs long.
Much of gene structure is broadly similar between eukaryotes and prokaryotes. These common elements largely result from the shared ancestry of cellular life in organisms over 2 billion years ago.[3] Key differences in gene structure between eukaryotes and prokaryotes reflect their divergent transcription and translation machinery.[4][5] Understanding gene structure is the foundation of understanding gene annotation, expression, and function.[6]
Previous images
Access to freely available diagrams is important for scientists, medical professions and the general public.[7][8] However, current open-access gene structure figures are limited in their scope (Supplementary figures 1-4), typically showing one or a few aspects (e.g. exon splicing, or promoter regions). Information-rich diagrams that use a consistent colour scheme and layout should help the comprehension of complex concepts.[9]
This work provides two diagrams that summarise the complex structure and terminology of genes. Common elements of gene structure are presented in a consistent layout and format to highlight the relationships between components. Key differences between eukaryotes and prokaryotes are indicated.
Results
Common gene structure features
The structures of both eukaryotic and prokaryotic genes involve several nested sequence elements. Each element has a specific function in the multi-step process of gene expression. The sequences and lengths of these elements vary, but the same general functions are present in most genes.[2] Although DNA is a double-stranded molecule, typically only one of the strands encodes information that the RNA polymerase reads to produce protein-coding mRNA or non-coding RNA. This 'sense' or 'coding' strand, runs in the 5' to 3' direction where the numbers refer to the carbon atoms of the backbone's ribose sugar. The open reading frame (ORF) of a gene is therefore usually represented as an arrow indicating the direction in which the sense strand is read.[10]
Regulatory sequences are located at the extremities of genes. These sequence regions can be next to the transcribed region (the promoter) or separated by many kilobases (enhancers and silencers).[11] The promoter is located at the 5' end of the gene and is composed of a core promoter sequence and a proximal promoter sequence. The core promoter marks the start site for transcription by binding RNA polymerase and other proteins necessary for copying DNA to RNA. The proximal promoter region binds transcription factors that modify the affinity of the core promoter for RNA polymerase.[12][13] Genes may be regulated by multiple enhancer and silencer sequences that further modify the activity of promoters by binding activator or repressor proteins.[14][15] Enhancers and silencers may be distantly located from the gene, many thousands of base pairs away. The binding of different transcription factors, therefore, regulates the rate of transcription initiation at different times and in different cells.[16]
Regulatory elements can overlap one another, with a section of DNA able to interact with many competing activators and repressors as well as RNA polymerase. For example, some repressor proteins can bind to the core promoter to prevent polymerase binding.[17] For genes with multiple regulatory sequences, the rate of transcription is the product of all of the elements combined.[18] Binding of activators and repressors to multiple regulatory sequences has a cooperative effect on transcription initiation.[19]
Although all organisms use both transcriptional activators and repressors, eukaryotic genes are said to be 'default off', whereas prokaryotic genes are 'default on'.[5] The core promoter of eukaryotic genes typically requires additional activation by promoter elements for expression to occur. The core promoter of prokaryotic genes, conversely, is sufficient for strong expression and is regulated by repressors.[5]
An additional layer of regulation occurs for protein coding genes after the mRNA has been processed to prepare it for translation to protein. Only the region between the start and stop codons encodes the final protein product. The flanking untranslated regions (UTRs) contain further regulatory sequences.[20] The 3' UTR contains a terminator sequence, which marks the endpoint for transcription and releases the RNA polymerase.[21] The 5’ UTR binds the ribosome, which translates the protein-coding region into a string of amino acids that fold to form the final protein product. In the case of genes for non-coding RNAs the RNA is not translated but instead folds to be directly functional.[22][23]
Eukaryotes

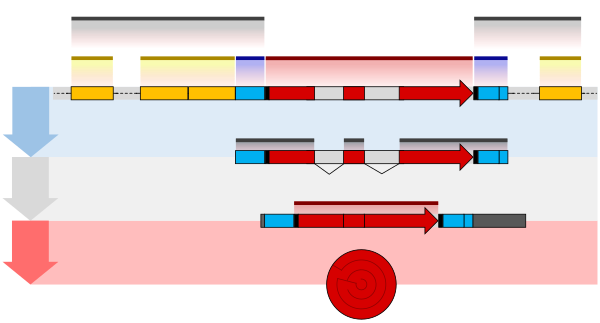
The structure of eukaryotic genes includes features not found in prokaryotes (Figure 1). Most of these relate to post-transcriptional modification of pre-mRNAs to produce mature mRNA ready for translation into protein. Eukaryotic genes typically have more regulatory elements to control gene expression compared to prokaryotes.[5] This is particularly true in multicellular eukaryotes, including humans, where gene expression varies widely among different tissues.[24]
A key feature of the structure of eukaryotic genes is that their transcripts are typically subdivided into exon and intron regions. Exon regions are retained in the final mature mRNA molecule, whereas intron regions are excised during post-transcriptional processing.[25] Indeed, the intron regions of a gene can be considerably longer than the exon regions. Once spliced together, the exons form a single continuous protein-coding region, and the splice boundaries are not detectable. Eukaryotic post-transcriptional processing also adds a 5' cap to the start of the mRNA and a poly-adenosine tail to the end of the mRNA. These additions stabilise the mRNA and direct its transport from the nucleus to the cytoplasm, although neither of these features are directly encoded in the structure of a gene.[20]
Prokaryotes

The overall organisation of prokaryotic genes is markedly different from that of the eukaryotes (Figure 2). The most obvious difference is that prokaryotic ORFs are often grouped into a polycistronic operon under the control of a shared set of regulatory sequences. These ORFs are all transcribed onto the same mRNA and so are co-regulated and often serve related functions.[26][27] Each ORF typically has its own ribosome binding site (RBS) so that ribosomes simultaneously translate ORFs on the same mRNA. Some operons also display translational coupling, where the translation rates of multiple ORFs within an operon are linked.[28] This can occur when the ribosome remains attached at the end of an ORF and simply translocates along to the next without the need for a new RBS.[29] Translational coupling is also observed when translation of an ORF affects the accessibility of the next RBS through changes in RNA secondary structure.[30] Having multiple ORFs on a single mRNA is only possible in prokaryotes because their transcription and translation take place at the same time and in the same subcellular location.[26][31]
The operator sequence next to the promoter is the main regulatory element in prokaryotes. Repressor proteins bound to the operator sequence physically obstruct the RNA polymerase enzyme, preventing transcription.[32][33] Riboswitches are other important regulatory sequences commonly present in prokaryotic UTRs. These sequences switch between alternative secondary structures in the RNA depending on the concentrations of key metabolites. The secondary structures then either block or reveal important sequence regions such as RBSs. Introns are extremely rare in prokaryotes and therefore do not play a significant role in prokaryotic gene regulation.[34]
Discussion
Strengths and limitations
The structures of genes are intimately linked to their functions. Genes are often thought of as only the protein-coding regions of DNA. However, coding sequences are only a minority component of the overall gene. Multiple untranscribed and untranslated DNA regions are necessary for proper gene function in all organisms. All regions of gene structure are important for determining phenotype, and so mutations in any of these sections have the potential to alter function.[35][36]
These two figures aim to present the key features of average genes. As such, they inevitably miss many interesting features, such as overlapping genes,[37] alternative splicing,[38] trans-splicing,[39] or the subcellular locations of events.[6] All the functions performed by the sequences shown are mediated through their interactions with pre-existing proteins and other trans-acting elements, which have been omitted for clarity. DNA is similarly stored in complex with histone proteins. Changes in this quaternary structure by epigenetic mechanisms such as histone modification or DNA methylation also affect how accessible genes are to the transcription machinery.[40] Despite these concessions, the diagrams provide an accurate overview of the important common aspects of gene structure necessary for understanding genetics.
Future extensions
These diagrams have been designed to be extended to other systems in a consistent visual format. For example, in addition to cellular gene structure, viruses have highly variable and complex genes with features dependent on their specific host. A diagram of gene features found in viruses would be of benefit to the community and could feature either model viruses, such as bacteriophage, or medically important examples, such as influenza or HIV.
RNA transcripts contain post-transcriptional modifications that are beyond the scope of the figures presented here. A figure outlining the RNA modifications that control transcript stability, transport, and translation would be a useful extension to these diagrams and extend the series from DNA into RNA structure. The transcriptional complex assembly and the spliceosomal complex assembly are two additional biological phenomena that would be well suited to this series. The protein components required and the order of assembly is well described yet the number of diagrams on Wikipedia is limited at this stage. Future works should extend the series by adding new concepts within the same unified format.
Conclusion
While these diagrams cover well established biological processes, there was an unmet need for logical diagrams with the clarity for readers to learn the basic concepts of gene structure. This work improves over existing representations by linking unifying concepts between eukaryotes and prokaryotes and explaining the concepts in a clear layout.
Availability
The diagrams are freely available in the following formats:
| Eukaryote | Prokaryote | |||
| Hyperlinked | Non-hyperlinked | Hyperlinked | Non-hyperlinked | |
| svg | link | link | link | link |
| Wikimarkup | link | - | link | - |
| png | - | link | - | link |
Unannotated versions for use in translations are available here and here.
Acknowledgements
The authors would like to thank Adrian J. Hunter, Boghog, and Opabinia regalis for their excellent recommendations during the Good Article review of Wikipedia's Gene page.
Declarations
Conflict of Interest: TS is a member of the Editorial Board of Wiki.J.Med. and has absented himself from all discussion and processing of this manuscript. Otherwise, we have no conflicts of interest.
References
- ↑ Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2002). "How Genetic Switches Work". Molecular Biology of the Cell (4 ed.). https://www.ncbi.nlm.nih.gov/books/NBK26872/.
- ↑ 2.0 2.1 2.2 Polyak, Kornelia; Meyerson, Matthew (2003). "Overview: Gene Structure". Cancer Medicine (6 ed.). BC Decker. https://www.ncbi.nlm.nih.gov/books/NBK12983/.
- ↑ Werner, Finn; Grohmann, Dina (2011). "Evolution of multisubunit RNA polymerases in the three domains of life". Nature Reviews Microbiology 9 (2): 85–98. doi:10.1038/nrmicro2507. ISSN 1740-1526.
- ↑ Kozak, Marilyn (1999). "Initiation of translation in prokaryotes and eukaryotes". Gene 234 (2): 187–208. doi:10.1016/S0378-1119(99)00210-3. ISSN 03781119.
- ↑ 5.0 5.1 5.2 5.3 Struhl, Kevin (1999). "Fundamentally Different Logic of Gene Regulation in Eukaryotes and Prokaryotes". Cell 98 (1): 1–4. doi:10.1016/S0092-8674(00)80599-1. ISSN 00928674.
- ↑ 6.0 6.1 Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2002). Molecular Biology of the Cell (Fourth ed.). New York: Garland Science. ISBN 978-0-8153-3218-3. http://www.ncbi.nlm.nih.gov/books/NBK21054/.
- ↑ Albert, Karen M. (2006-07-01). "Open access: implications for scholarly publishing and medical libraries". Journal of the Medical Library Association: JMLA 94 (3): 253–262. ISSN 1558-9439. PMID 16888657. https://www.ncbi.nlm.nih.gov/pubmed/16888657.
- ↑ Masukume, G; Kipersztok, L; Das, D; Shafee, T; Laurent, M; Heilman, J (November 2016). "Medical journals and Wikipedia: a global health matter". The Lancet Global Health 4 (11): e791. doi:10.1016/S2214-109X(16)30254-6.
- ↑ Rougier, Nicolas P.; Droettboom, Michael; Bourne, Philip E. (2014-09-11). "Ten Simple Rules for Better Figures". PLOS Comput Biol 10 (9): e1003833. doi:10.1371/journal.pcbi.1003833. ISSN 1553-7358. PMID 25210732. PMC 4161295. http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1003833.
- ↑ Lu, G. (2004). "Vector NTI, a balanced all-in-one sequence analysis suite". Briefings in Bioinformatics 5 (4): 378–388. doi:10.1093/bib/5.4.378. ISSN 1467-5463.
- ↑ Wiper-Bergeron, Nadine; Skerjanc, Ilona S. (2009). Transcription and the Control of Gene Expression. Humana Press. pp. 33–49. doi:10.1007/978-1-59745-440-7_2. ISBN 978-1-59745-440-7.
- ↑ Thomas, Mary C.; Chiang, Cheng-Ming (2008). "The General Transcription Machinery and General Cofactors". Critical Reviews in Biochemistry and Molecular Biology 41 (3): 105–178. doi:10.1080/10409230600648736. ISSN 1040-9238.
- ↑ Juven-Gershon, Tamar; Hsu, Jer-Yuan; Theisen, Joshua WM; Kadonaga, James T (2008). "The RNA polymerase II core promoter — the gateway to transcription". Current Opinion in Cell Biology 20 (3): 253–259. doi:10.1016/j.ceb.2008.03.003. ISSN 09550674.
- ↑ Maston, Glenn A.; Evans, Sara K.; Green, Michael R. (2006). "Transcriptional Regulatory Elements in the Human Genome". Annual Review of Genomics and Human Genetics 7 (1): 29–59. doi:10.1146/annurev.genom.7.080505.115623. ISSN 1527-8204.
- ↑ Pennacchio, L. A.; Bickmore, W.; Dean, A.; Nobrega, M. A.; Bejerano, G. (2013). "Enhancers: Five essential questions". Nature Reviews Genetics 14 (4): 288–95. doi:10.1038/nrg3458. PMID 23503198.
- ↑ Maston, G. A.; Evans, S. K.; Green, M. R. (2006). "Transcriptional Regulatory Elements in the Human Genome". Annual Review of Genomics and Human Genetics 7: 29–59. doi:10.1146/annurev.genom.7.080505.115623. PMID 16719718.
- ↑ Ogbourne, Steven; Antalis, Toni M. (1998). "Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes". Biochemical Journal 331 (1): 1–14. doi:10.1042/bj3310001. ISSN 0264-6021.
- ↑ Buchler, N. E.; Gerland, U.; Hwa, T. (2003). "On schemes of combinatorial transcription logic". Proceedings of the National Academy of Sciences 100 (9): 5136–5141. doi:10.1073/pnas.0930314100. ISSN 0027-8424.
- ↑ Kazemian, M.; Pham, H.; Wolfe, S. A.; Brodsky, M. H.; Sinha, S. (11 July 2013). "Widespread evidence of cooperative DNA binding by transcription factors in Drosophila development". Nucleic Acids Research 41 (17): 8237–8252. doi:10.1093/nar/gkt598.
- ↑ 20.0 20.1 Guhaniyogi, Jayita; Brewer, Gary (2001). "Regulation of mRNA stability in mammalian cells". Gene 265 (1-2): 11–23. doi:10.1016/S0378-1119(01)00350-X. ISSN 03781119.
- ↑ Kuehner, Jason N.; Pearson, Erika L.; Moore, Claire (2011). "Unravelling the means to an end: RNA polymerase II transcription termination". Nature Reviews Molecular Cell Biology 12 (5): 283–294. doi:10.1038/nrm3098. ISSN 1471-0072.
- ↑ Mattick, J. S. (2006). "Non-coding RNA". Human Molecular Genetics 15 (90001): R17–R29. doi:10.1093/hmg/ddl046. ISSN 0964-6906.
- ↑ Palazzo, Alexander F.; Lee, Eliza S. (2015). "Non-coding RNA: what is functional and what is junk?". Frontiers in Genetics 6. doi:10.3389/fgene.2015.00002. ISSN 1664-8021.
- ↑ Maston, Glenn A.; Evans, Sara K.; Green, Michael R. (2006). "Transcriptional Regulatory Elements in the Human Genome". Annual Review of Genomics and Human Genetics 7 (1): 29–59. doi:10.1146/annurev.genom.7.080505.115623. ISSN 1527-8204.
- ↑ Matera, A. Gregory; Wang, Zefeng (2014). "A day in the life of the spliceosome". Nature Reviews Molecular Cell Biology 15 (2): 108–121. doi:10.1038/nrm3742. ISSN 1471-0072.
- ↑ 26.0 26.1 Salgado, H.; Moreno-Hagelsieb, G.; Smith, T.; Collado-Vides, J. (2000). "Operons in Escherichia coli: Genomic analyses and predictions". Proceedings of the National Academy of Sciences 97 (12): 6652–6657. doi:10.1073/pnas.110147297. PMID 10823905. PMC 18690. //www.ncbi.nlm.nih.gov/pmc/articles/PMC18690/.
- ↑ Jacob, F.; Monod, J. (1961-06-01). "Genetic regulatory mechanisms in the synthesis of proteins". Journal of Molecular Biology 3: 318–356. doi:10.1016/S0022-2836(61)80072-7. ISSN 0022-2836. PMID 13718526. https://www.ncbi.nlm.nih.gov/pubmed/13718526.
- ↑ Tian, Tian; Salis, Howard M. (2015). "A predictive biophysical model of translational coupling to coordinate and control protein expression in bacterial operons". Nucleic Acids Research 43 (14): 7137–7151. doi:10.1093/nar/gkv635. ISSN 0305-1048.
- ↑ Schümperli, Daniel; McKenney, Keith; Sobieski, Donna A.; Rosenberg, Martin (1982). "Translational coupling at an intercistronic boundary of the Escherichia coli galactose operon". Cell 30 (3): 865–871. doi:10.1016/0092-8674(82)90291-4. ISSN 00928674.
- ↑ Levin-Karp, Ayelet; Barenholz, Uri; Bareia, Tasneem; Dayagi, Michal; Zelcbuch, Lior; Antonovsky, Niv; Noor, Elad; Milo, Ron (2013). "Quantifying Translational Coupling in E. coli Synthetic Operons Using RBS Modulation and Fluorescent Reporters". ACS Synthetic Biology 2 (6): 327–336. doi:10.1021/sb400002n. ISSN 2161-5063.
- ↑ Lewis, Mitchell (June 2005). "The lac repressor". Comptes Rendus Biologies 328 (6): 521–548. doi:10.1016/j.crvi.2005.04.004. http://www.sciencedirect.com/science/article/pii/S1631069105000685.
- ↑ McClure, W R (1985). "Mechanism and Control of Transcription Initiation in Prokaryotes". Annual Review of Biochemistry 54 (1): 171–204. doi:10.1146/annurev.bi.54.070185.001131. ISSN 0066-4154.
- ↑ Bell, Charles E; Lewis, Mitchell (2001). "The Lac repressor: a second generation of structural and functional studies". Current Opinion in Structural Biology 11 (1): 19–25. doi:10.1016/S0959-440X(00)00180-9. ISSN 0959440X.
- ↑ Rodríguez-Trelles, Francisco; Tarrío, Rosa; Ayala, Francisco J. (2006). "Origins and Evolution of Spliceosomal Introns". Annual Review of Genetics 40 (1): 47–76. doi:10.1146/annurev.genet.40.110405.090625. ISSN 0066-4197.
- ↑ Wray, G. A. (2003). "The Evolution of Transcriptional Regulation in Eukaryotes". Molecular Biology and Evolution 20 (9): 1377–1419. doi:10.1093/molbev/msg140. ISSN 0737-4038.
- ↑ Wray, Gregory A. (2007). "The evolutionary significance of cis-regulatory mutations". Nature Reviews Genetics 8 (3): 206–216. doi:10.1038/nrg2063. ISSN 1471-0056.
- ↑ Krakauer, David C. (2000). "Stability and Evolutionof Overlapping Genes". Evolution 54 (3): 731–739. doi:10.1111/j.0014-3820.2000.tb00075.x. ISSN 0014-3820.
- ↑ Kornblihtt, Alberto R.; Schor, Ignacio E.; Alló, Mariano; Dujardin, Gwendal; Petrillo, Ezequiel; Muñoz, Manuel J. (2013). "Alternative splicing: a pivotal step between eukaryotic transcription and translation". Nature Reviews Molecular Cell Biology 14 (3): 153–165. doi:10.1038/nrm3525. ISSN 1471-0072.
- ↑ Yang, Y; Walsh, C (2005). "Spliceosome-Mediated RNA -splicing". Molecular Therapy 12 (6): 1006–1012. doi:10.1016/j.ymthe.2005.09.006. ISSN 15250016.
- ↑ Mazzio, Elizabeth A.; Soliman, Karam F.A. (2014). "Basic concepts of epigenetics". Epigenetics 7 (2): 119–130. doi:10.4161/epi.7.2.18764. ISSN 1559-2294.
Supplementary figures
A simple bar example of intron and exon arrangement
David. D, CC-BY-SA